Tutorial for the Neo4j backend¶
Part 2: Rewriting hierarchies of graphs¶
ReGraph allows to create a hierarchies of graphs related by means of homomorphisms (or typing). In the context of a hierarchy, if there exists a homomorphism G -> T, we say that graph G is typed by a graph T. Graph hierarchy is a DAG, where nodes are graphs and edges are homomorphisms. A homomorphism maps every node of G to some node in T (a type) in such a way that:
edges are preserved
attributes of both nodes and edges are preserved
ReGraph provides the data structure Neo4jHierarchy that encodes a graph hierarchy as a property graph stored in a Neo4j database. This encoding represents each graph in a hierarchy with nodes of the database labeled by a unique label corresponding to the ID of the corresponding graph in the hierarchy. For example, the following Cypher query mathces all the nodes belonging to the graph graphID in the hierarchy:
MATCH (n:graphID) RETURN n
The edges in graphs are labeled as edge and the typing edges are labeled as typing. We can then easily find the typing of nodes in graph1 by graph2 with the query:
MATCH (n:graph1)-[:typing]->(m:graph2) RETURN n, m
The hierarchy skeleton is represented by nodes labeled as graph be default. So, matching the nodes of the hierarchy skeleton can be done with the following query:
MATCH (n:graph) return n
Let us start by importing the necessary data structures and some utilities:
from regraph import NXGraph, Neo4jHierarchy, Rule
from regraph import plot_graph, plot_instance, plot_rule
Creating and modifying hierarchies¶
Consider the following example of a simple graph hierarchy. The two graphs G and T are being created and added to the heirarchy. After, a typing homomorphism between G and T is added, so that every node of G is typed by some node in T.
# Define graph G
g = NXGraph()
g.add_nodes_from(["protein", "binding", "region", "compound"])
g.add_edges_from([
("region", "protein"),
("protein", "binding"),
("region", "binding"),
("compound", "binding")])
# Create a hierarchy
simple_hierarchy = Neo4jHierarchy(
uri="bolt://localhost:7687",
user="neo4j", password="neo4j")
simple_hierarchy.add_graph(
"G", g, {"name": "Simple protein interaction"})
# Add a new graph without explicitly creating a graph object
simple_hierarchy.add_graph_from_data(
"T",
node_list=["action", "agent"],
edge_list=[("agent", "agent"), ("agent", "action")],
attrs={"name": "Agent interaction"})
simple_hierarchy.add_typing(
"G", "T",
{"protein": "agent",
"region": "agent",
"compound": "agent",
"binding": "action",
}
)
The method get_graph returns the graph object corresponding to the provided graph id.
>>> type(simple_hierarchy.get_graph("T"))
regraph.backends.neo4j.graphs.Neo4jGraph
The method get_typing returns the dictionary object corresponding to the provided hierarchy edge and representing the associated graph homomorphism.
>>> simple_hierarchy.get_typing("G", "T")
{'protein': 'agent',
'region': 'agent',
'compound': 'agent',
'binding': 'action'}
Rewriting of objects in a hierarchy¶
ReGraph implements the rewriting technique called sesqui-pushout rewriting that allows to transform graphs by applying rules through their instances (matchings). Rewriting an individual graphs in a hierarchy may require an update of other graphs and typings in this hierarchy, such updates are called propagation and are distinguished into two types: backward and forward propagation.
Backward propagation briefly: - If some graph elements (nodes/edges or attributes) are removed from a graph in the hierarchy, then all the respective elements that are typed by them in the ancestor graphs should be removed. - If a graph node is cloned, then for every instance of this node (every node that is typed by the clonned node) in the ancestor graphs we either: (a) specify to which clone it corresponds or (b) clone it.
Forward propagation briefly: - If some graph nodes are merged and these nodes are typed by different nodes in a descendant graph, the corresponding nodes in the descendant graph should be merged. - If a new graph element (node/edge or attribute) is added, then for all the descendent graphs in the hierarchy we either (a) select an existing element to type the added element or (b) add a new element to type the added element.
For more details, please see here.
ReGraph allows to rewrite individual graphs situated in the hierarchy using the method rewrite of NXHierarchy. The rewriting can be done in two modes:
Strict rewriting rewriting that does not allow propagation.
Non-strict rewriting that allows propagation.
The rewrite takes as the input the following parameters:
graph_id, ID of the graph in the hierarchy to rewrite,
rule, a rule object to apply,
instance, a dictionary containing an instance of the lhs of the rule in the graph subject to rewriting, by default, tries to construct identity morphism of the nodes of the pattern,
p_typing, a dictionary containing typing of graphs in the hierarchy by the interface of the rule, keys are IDs of hierarchy graphs, values are dictionaries containing the mapping of nodes from the hierarchy graphs to the inteface nodes (note that a node from a graph can be typed by a set of nodes in the interface of the rule, e.g. if we want to perform cloning of some types, etc).
rhs_typing, a dictionary containing typing of the rhs by graphs of the hierarchy, keys are ids of hierarchy graphs, values are dictionaries containing the mapping of nodes from the lhs to the nodes of the typing graph given by the respective key of the value (note that a node from the rhs can be typed by a set of nodes of some graph, e.g. if we want to perform merging of some types, etc),
strict, flag indicating if rewriting is strict, then any propagation is not allowed.
Strict rewriting¶
Let us create a Rule object containing a rule we would like to apply.
lhs = NXGraph()
lhs.add_nodes_from([1, 2])
lhs.add_edges_from([(1, 2)])
p = NXGraph()
p.add_nodes_from([1, 2])
p.add_edges_from([])
rhs = NXGraph()
rhs.add_nodes_from([1, 2, 3])
rhs.add_edges_from([(3, 1), (3, 2)])
# By default if `p_lhs` and `p_rhs` are not provided
# to a rule, it tries to construct this homomorphisms
# automatically by matching the names. In this case we
# have defined lhs, p and rhs in such a way that that
# the names of the matching nodes correspond
rule = Rule(p, lhs, rhs)
plot_rule(rule)

The created rule removes the edge 1->2, adds the new node 3 and two edges 3->1 and 3->2. Let us find instances of the created rule in the graph G as follows:
>>> instances = simple_hierarchy.find_matching("G", rule.lhs)
>>> print("Instances: ", instances)
Instances: [{'a': 'protein', 'b': 'binding'}, {'a': 'compound', 'b': 'binding'}, {'a': 'region', 'b': 'binding'}, {'a': 'region', 'b': 'protein'}]
Let us fix the desired instance: we would like to remove the edge from protein to binding and add some new node connecting them.
>>> instance = {1: "protein", 2: "binding"}
Let us try to apply the rule to the selected instance as is in the strict rewriting mode:
try:
rhs_instance = simple_hierarchy.rewrite("G", rule, instance, strict=True)
except Exception as e:
print("Error message: ", e)
print("Type: ", type(e))
Running this snippet produces the following output:
Error message: Rewriting is strict (no propagation of types is allowed), typing of the added nodes '{3}' by 'T' is required
Type: <class 'regraph.exceptions.RewritingError'>
We have failed to rewrite G, because we have not specified typing for the newly added node 3. Let us try again, but this time we will prove such typing.
rhs_typing = {"T": {3: "agent"}}
rhs_instance = simple_hierarchy.rewrite(
"G", rule, instance, rhs_typing=rhs_typing, strict=True)
We can check the instance of the right-hand side of the rule:
>>> print(rhs_instance)
{'a': 'protein', 'b': 'binding', 'c': 'c'}
Let us now create a rule that applied to T and that clones the node agent into two nodes.
lhs = NXGraph()
lhs.add_nodes_from(["agent"])
rule = Rule.from_transform(lhs)
_, rhs_clone = rule.inject_clone_node("agent")
plot_rule(rule)

We set its instance explicitly:
>>> instance = {"agent": "agent"}
and we try to apply the created rule to the graph T in the strict mode:
try:
rhs_instance = simple_hierarchy.rewrite("T", rule, instance, strict=True)
except Exception as e:
print("Error message: ", e)
print("Type: ", type(e))
Running this snippet produces the following output:
Error message: Rewriting is strict (no propagation of clones is allowed), the cloned node 'agent' in 'T' has instances '['protein', 'region', 'compound', 3]' in 'G' and their typing by P is not specified
Type: <class 'regraph.exceptions.RewritingError'>
We have failed to rewrite T, because we have not specified typing for instances of agent in the interface of the rule (P). Let us try again, but this time we will prove such typing:
p_typing = {
"G": {
'protein': 'agent',
'region': 'agent',
'compound': rhs_clone,
3: 'agent'
}
}
rhs_instance = simple_hierarchy.rewrite(
"T", rule, instance, p_typing=p_typing, strict=True)
Let us relabel nodes in T.
>>> simple_hierarchy.relabel_graph_node('T', rhs_instance['agent'], 'organic_agent')
>>> simple_hierarchy.relabel_graph_node('T', rhs_instance[rhs_clone], 'non_organic_agent')
>>> plot_graph(simple_hierarchy.get_graph('T'))
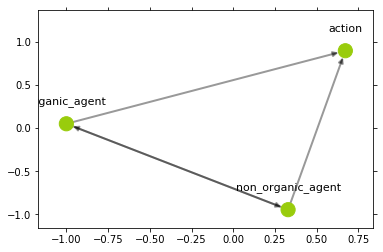
>>> print(simple_hierarchy.get_typing("G", "T"))
{'protein': 'organic_agent', 'binding': 'action', 'region': 'organic_agent', 'compound': 'non_organic_agent', 3: 'organic_agent'}
Rewriting and propagation¶
To illustrate rewriting with propagation, let us consider a slighlty more sophisticated hierarchy. The following snippet creates a new Neo4jHierarchy object.
hierarchy = Neo4jHierarchy(
uri="bolt://localhost:7687",
user="neo4j",
password="neo4j")
The new hierarchy object provides connection to the Neo4j database. If you have run the previous example, hierarchy will connect to the same database as the previously used simple_graph and will contain the same graphs G and T. You can call the following method to clear the database before proceeding:
>>> hierarchy._clear()
Let us add graphs and homomorphisms to the new hierarchy:
colors = NXGraph()
colors.add_nodes_from([
"green", "red"
])
colors.add_edges_from([
("red", "green"),
("red", "red"),
("green", "green")
])
hierarchy.add_graph("colors", colors)
shapes = NXGraph()
shapes.add_nodes_from(["circle", "square"])
shapes.add_edges_from([
("circle", "square"),
("square", "circle"),
("circle", "circle")
])
hierarchy.add_graph("shapes", shapes)
quality = NXGraph()
quality.add_nodes_from(["good", "bad"])
quality.add_edges_from([
("bad", "bad"),
("bad", "good"),
("good", "good")
])
hierarchy.add_graph("quality", quality)
g1 = NXGraph()
g1.add_nodes_from([
"red_circle",
"red_square",
])
g1.add_edges_from([
("red_circle", "red_square"),
("red_circle", "red_circle"),
("red_square", "red_circle")
])
g1_colors = {
"red_circle": "red",
"red_square": "red",
}
g1_shapes = {
"red_circle": "circle",
"red_square": "square",
}
hierarchy.add_graph("g1", g1)
hierarchy.add_typing("g1", "colors", g1_colors)
hierarchy.add_typing("g1", "shapes", g1_shapes)
g2 = NXGraph()
g2.add_nodes_from([
"good_circle",
"good_square",
"bad_circle",
])
g2.add_edges_from([
("good_circle", "good_square"),
("good_square", "good_circle"),
("bad_circle", "good_circle"),
("bad_circle", "bad_circle"),
])
g2_shapes = {
"good_circle": "circle",
"good_square": "square",
"bad_circle": "circle"
}
g2_quality = {
"good_circle": "good",
"good_square": "good",
"bad_circle": "bad",
}
hierarchy.add_graph("g2", g2)
hierarchy.add_typing("g2", "shapes", g2_shapes)
hierarchy.add_typing("g2", "quality", g2_quality)
g3 = NXGraph()
g3.add_nodes_from([
"good_red_circle",
"bad_red_circle",
"good_red_square",
])
g3.add_edges_from([
("bad_red_circle", "good_red_circle"),
("good_red_square", "good_red_circle"),
("good_red_circle", "good_red_square")
])
g3_g1 = {
"good_red_circle": "red_circle",
"bad_red_circle": "red_circle",
"good_red_square": "red_square"
}
g3_g2 = {
"good_red_circle": "good_circle",
"bad_red_circle": "bad_circle",
"good_red_square": "good_square",
}
hierarchy.add_graph("g3", g3)
hierarchy.add_typing("g3", "g1", g3_g1)
hierarchy.add_typing("g3", "g2", g3_g2)
Some of the graphs in the hierarchy are now typed by multiple graphs, which is reflected in the types of nodes, as in the example below:
print("Node types in G3:\n")
for node in hierarchy.get_graph("g3").nodes():
print(node, hierarchy.node_type("g3", node))
outputs:
Node types in G3:
good_red_circle {'g1': 'red_circle', 'g2': 'good_circle'}
bad_red_circle {'g1': 'red_circle', 'g2': 'bad_circle'}
good_red_square {'g1': 'red_square', 'g2': 'good_square'}
We now show how graph rewriting can be performed in such an hierarchy. In the previous example we perfromed strict rewriting in a hierarchy, where no propagation was performed.
The following example illustrates how the ReGraph propagates the changes made by rewriting on any level to all the graphs (as well as the rules) typed by the one target of rewriting.
lhs = NXGraph()
lhs.add_nodes_from(["a", "b"])
lhs.add_edges_from([
("a", "b"),
("b", "a")
])
p = NXGraph()
p.add_nodes_from(["a", "a1", "b"])
p.add_edges_from([
("a", "b"),
("a1", "b")
])
rhs = NXGraph.copy(p)
rule = Rule(
p, lhs, rhs,
{"a": "a", "a1": "a", "b": "b"},
{"a": "a", "a1": "a1", "b": "b"},
)
plot_rule(rule)
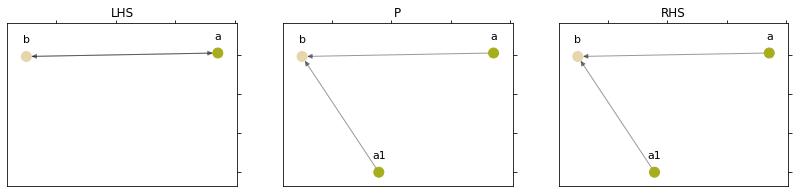
We have created a rule that clones the node a and reconnects the edges between a and b. We rewrite the graph shapes with the fixed instances (so that the node circle is cloned).
>>> rhs_instances = hierarchy.rewrite(
>>> "shapes", rule, {"a": "circle", "b": "square"})
Observe the following snippets, the cloning of circle was propagated to all the ancestors of shapes, because we didn’t specify how to retype intances of circle for these ancestors using the p_typing parameter. This is an example of previously mentioned backward propagation.
>>> print(hierarchy.get_graph("shapes").nodes())
['circle', 'square', 'circle1']
>>> print(hierarchy.get_graph("g1").nodes())
['red_circle', 'red_square', 'red_circle1']
>>> print(hierarchy.get_graph("g2").nodes())
['good_circle', 'good_square', 'bad_circle', 'good_circle1', 'bad_circle1']
>>> print(hierarchy.get_graph("g3").nodes())
['good_red_circle2', 'bad_red_circle2', 'good_red_circle11', 'bad_red_circle11', 'good_red_circle', 'bad_red_circle', 'good_red_square', 'good_red_circle1', 'bad_red_circle1']
Let us now consider a small example of forward propagation. We will create a rule that performs some additions and merges of nodes.
pattern = NXGraph()
pattern.add_nodes_from(["a", "b"])
rule = Rule.from_transform(pattern)
rhs_node = rule.inject_merge_nodes(["a", "b"])
rule.inject_add_node("c")
rule.inject_add_edge("c", rhs_node)
instance = {
"a": "good_circle",
"b": "bad_circle",
}
plot_rule(rule)

Application of this rule will merge nodes bad_circle and good_circle in the graph g2. It with then add a new node and connect it with an edge to the merged node. Let us specify some typings of the new node in the RHS: we will set the new node to be typed as circle in the graph shapes.
rhs_typing = {
"shapes": {
"c": "circle"
}
}
rhs_instance = hierarchy.rewrite(
"g2", rule, instance, rhs_typing=rhs_typing)
Observe the following snippets, as the result of forward propagation nodes good and bad were merged in the graph quality. In addition, a new node typing the node c in the rule was added to the graph quality.
>>> print(rhs_instance)
{'a_b': 'bad_circle_good_circle', 'c': 'c'}
>>> print(hierarchy.get_graph("quality").nodes())
['bad_good', 'c']
Serializing hierarchy objects¶
ReGraph provides some utils for serialization of Neo4jHierarchy objects and implements the following methods for loading and exporting your hierarchy in JSON-format:
Neo4jHierarchy.to_json creates a json representations of the hierarchy;
Neo4jHierarchy.from_json loads an hierarchy from json representation (returns new Hierarchy object);
Neo4jHierarchy.export exports the hierarchy to a file (json format);
Neo4jHierarchy.load loads an hierarchy from a .json file (returns new object as well).
hierarchy_json = hierarchy.to_json()
# Clear the DB for the previous hierarchy
hierarchy._clear()
# Load json-data back to the DB
hierarchy = Neo4jHierarchy.from_json(
uri="bolt://localhost:7687", user="neo4j",
password="neo4j", json_data=hierarchy_json)
The presented snippet creates a new hierarchy object by loading the JSON-serialized data into the provided Neo4j database.
Next¶
Continue to Part 3: Rewriting schema-aware PGs through TypedNeo4jGraph rewriting schema-aware PGs.
See more¶
Module reference: Neo4j-based hierarchies